The title of this post comes from the site www.crossref.org/members/prep/ Here you can explore how your favourite publisher of scientific articles exposes metadata for their journal.
Firstly, a reminder that when an article is published, the publisher collects information about the article (the “metadata”) and registers this information with CrossRef in exchange for a DOI. This metadata in turn is used to power e.g. a search engine which allows “rich” or “deep” searching of the articles to be undertaken. There is also what is called an API (Application Programmer Interface) which allows services to be built offering deeper insights into what are referred to as scientific objects. One such service is “Event Data“, which attempts to create links between various research objects such as publications, citations, data and even commentaries in social media. A live feed can be seen here.
So here are the results for the metadata provided by six publishers familiar to most chemists, with categories including;
- References
- Open References
- ORCID IDs
- Text mining URLs
- Abstracts
One immediately notices the large differences between publishers. Thus most have 0% metadata for the article abstracts, but one (the RSC) has 87%! Another striking difference is those that support open references (OpenCitations). The RSC and Springer Nature are 99-100% compliant whilst the ACS is 0%. Yet another variation is the adoption of the ORCID (Open Researcher and Collaborator Identifier), where the learned society publishers (RSC, ACS) achieve > 80%, but the commercial publishers are in the lower range of 20-49%.
To me the most intriguing was the Text mining URLs. From the help pages, “The Crossref REST API can be used by researchers to locate the full text of content across publisher sites. Publishers register these URLs – often including multiple links for different formats such as PDF or XML – and researchers can request them programatically“. Here the RSC is at 0%, ACS is at 8% but the commercial publishers are 80+%. I tried to find out more at e.g. https://www.springernature.com/gp/researchers/text-and-data-mining but the site was down when I tried. This can be quite a controversial area. Sometimes the publisher exerts strict control over how the text mining can be carried out and how any results can be disseminated. Aaron Swartz famously fell foul of this.
I am intrigued as to how, as a reader with no particular pre-assembled toolkit for text mining, I can use this metadata provided by the publishers to enhance my science. After all, 80+% of articles with some of the publishers apparently have a mining URL that I could use programmatically. If anyone reading this can send some examples of the process, I would be very grateful.
Finally I note the absence of any metadata in the above categories relating to FAIR data. Such data also has the potential for programmatic procedures to retrieve and re-use it (some examples are available here[1]), but apparently publishers do not (yet) collect metadata relating to FAIR. Hopefully they soon will.
References
- A. Barba, S. Dominguez, C. Cobas, D.P. Martinsen, C. Romain, H.S. Rzepa, and F. Seoane, "Workflows Allowing Creation of Journal Article Supporting Information and Findable, Accessible, Interoperable, and Reusable (FAIR)-Enabled Publication of Spectroscopic Data", ACS Omega, vol. 4, pp. 3280-3286, 2019. https://doi.org/10.1021/acsomega.8b03005
Tags: Aaron Swartz, Academic publishing, API, Business intelligence, CrossRef, data, Data management, Elsevier, favourite publisher, Identifiers, Information, Information science, Knowledge, Knowledge representation, metadata, mining, ORCiD, PDF, Pre-exposure prophylaxis, Publishing, Publishing Requirements for Industry Standard Metadata, Records management, Research Object, Scholarly communication, Scientific literature, search engine, social media, Technical communication, Technology/Internet, text mining, Written communication, XML
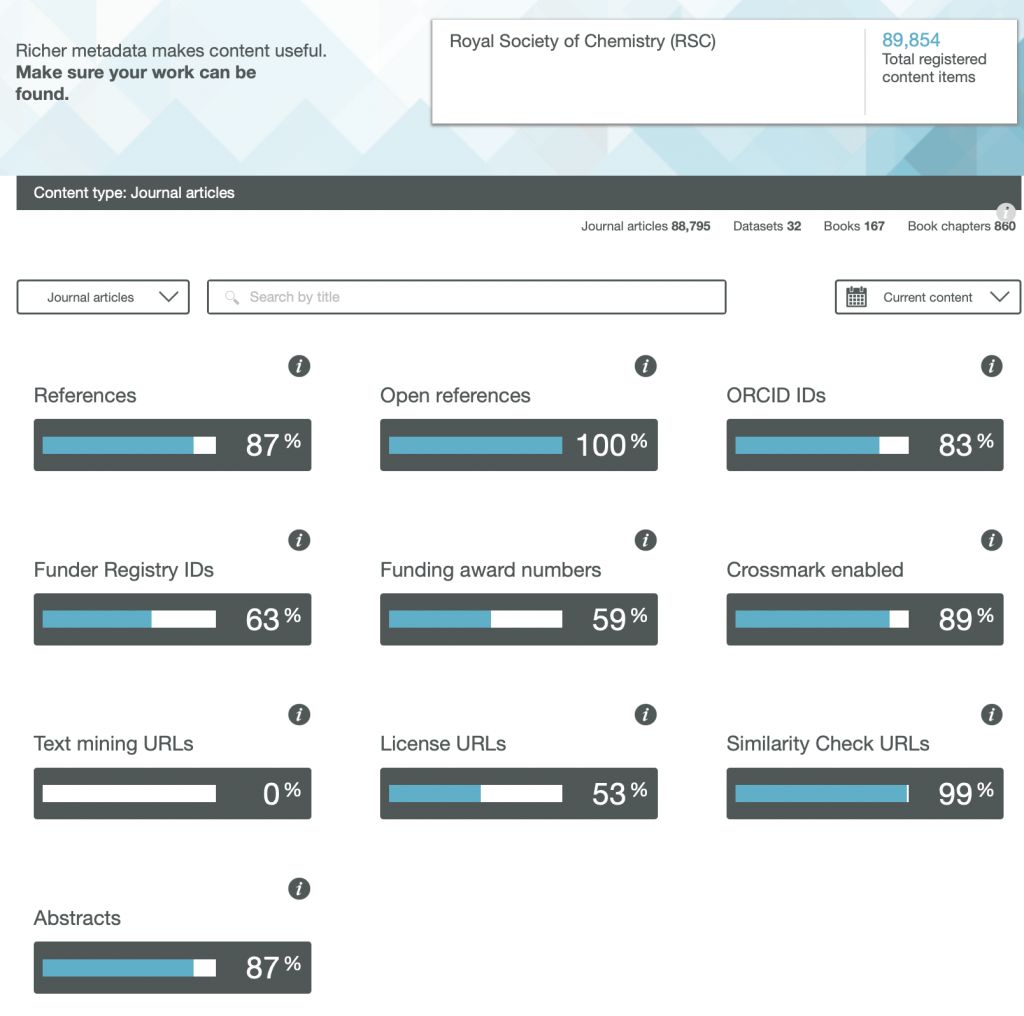
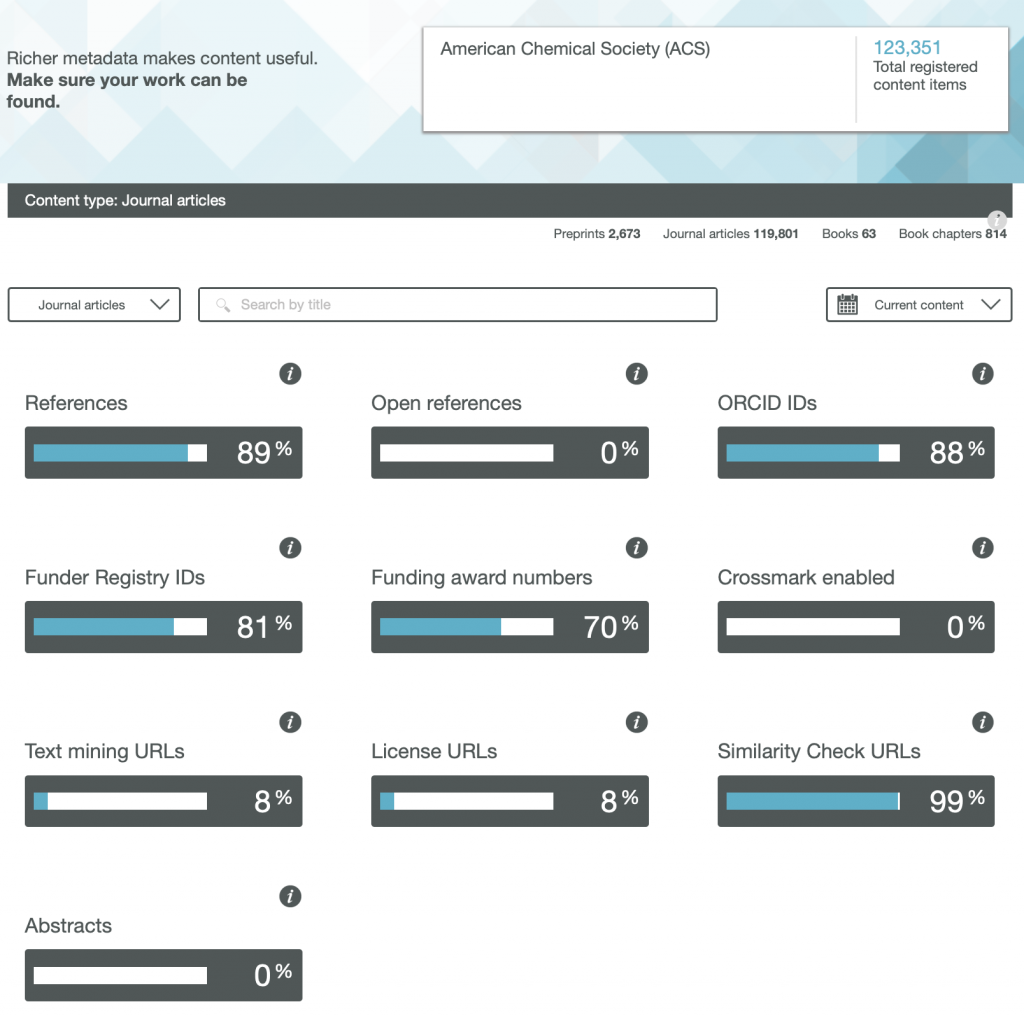
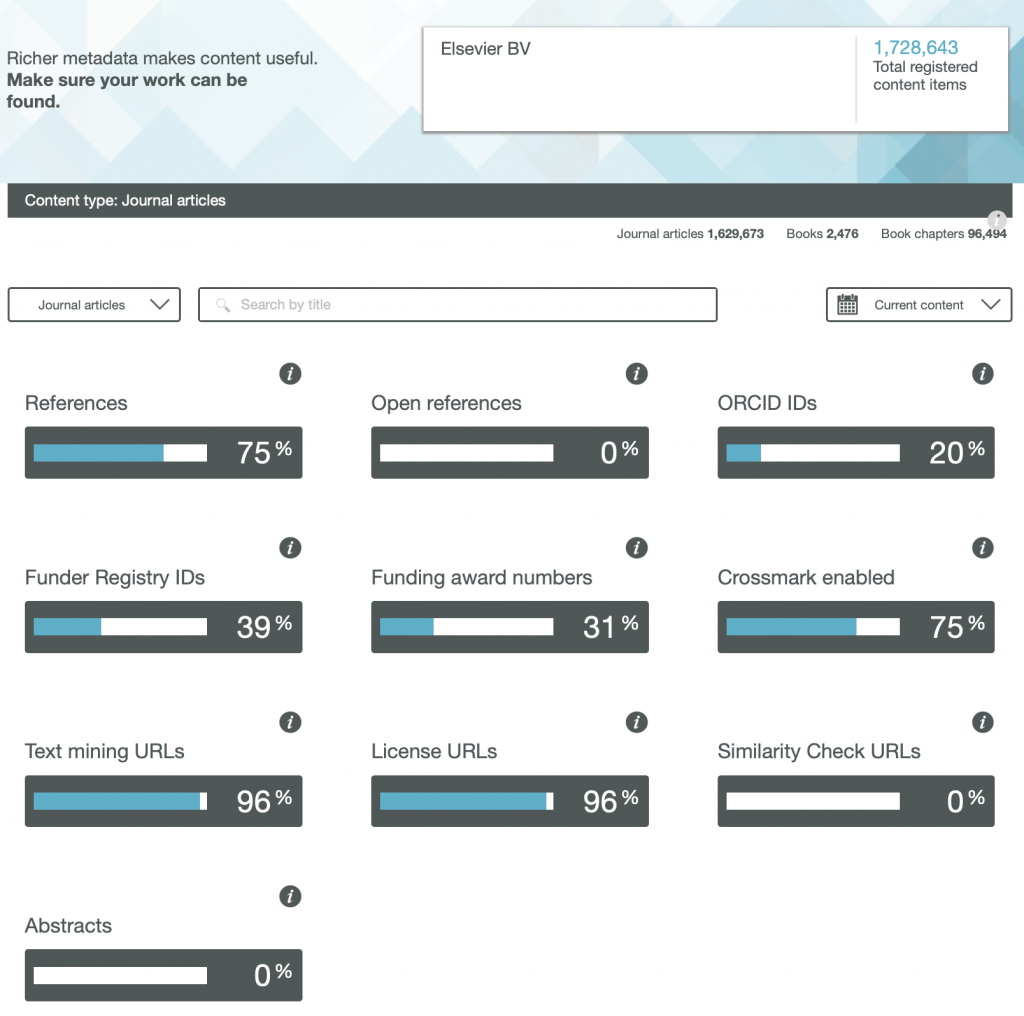
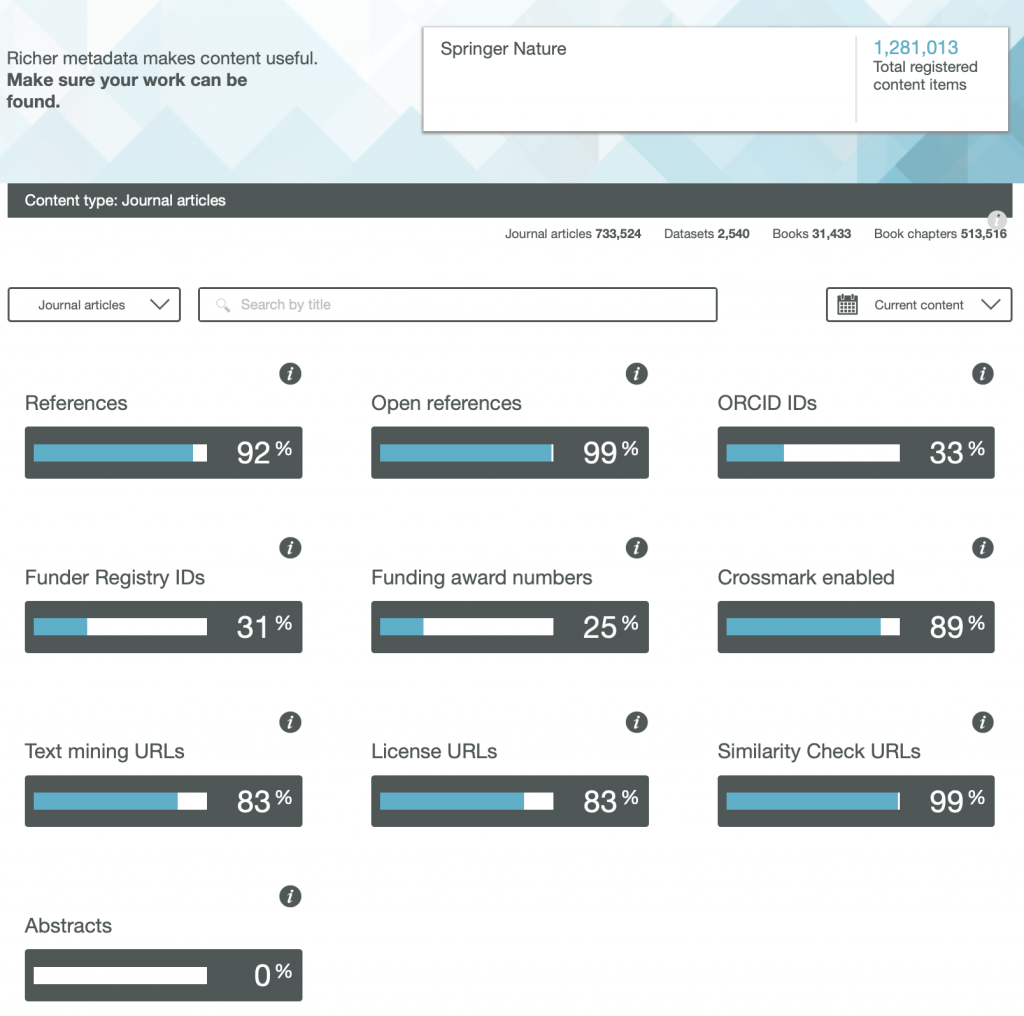
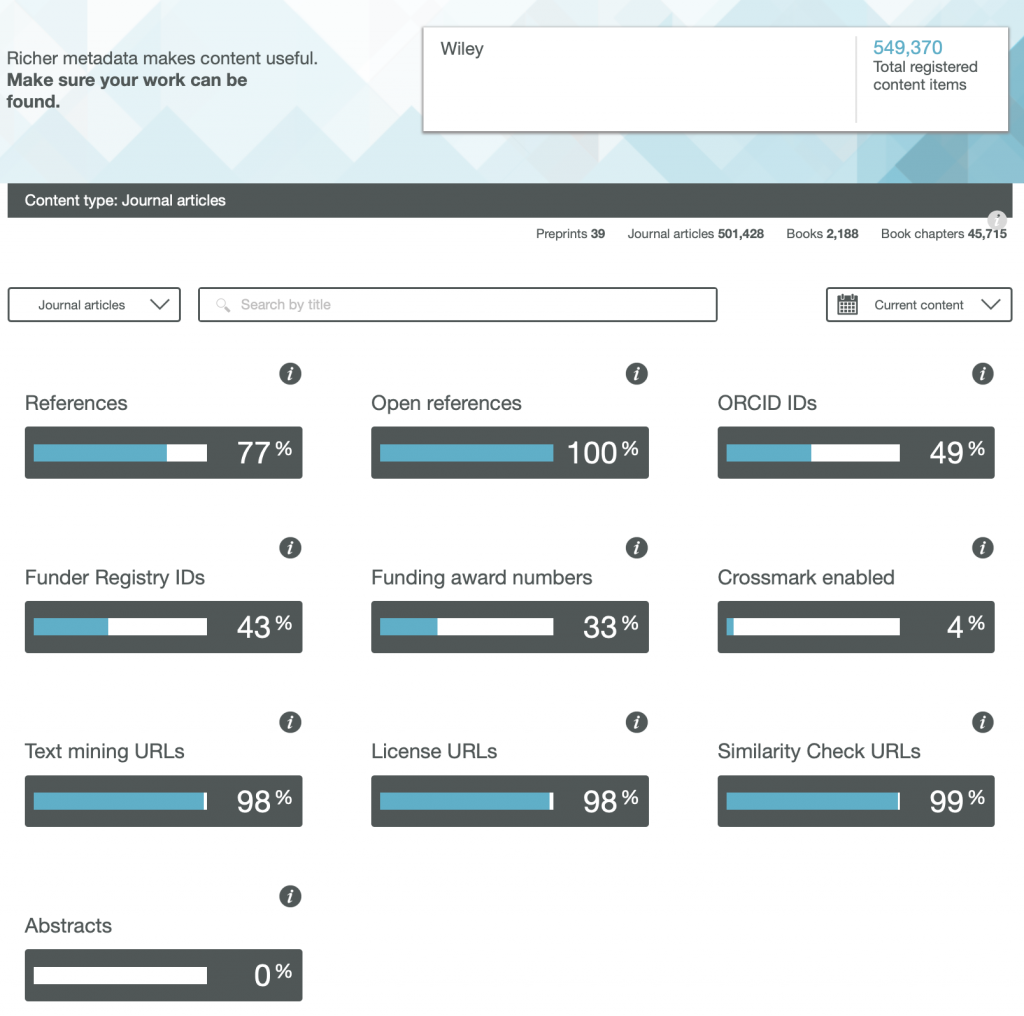
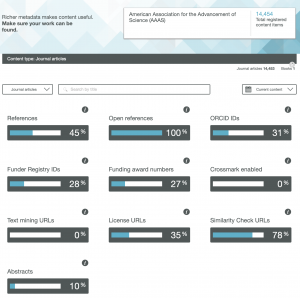
Ted Habermann has analysed the metadata statistics for several journals using the CrossRef API. The results are here: https://www.tedhabermann.com/blog/2019/2/19/metadata-evolution-crossref-participation-reports